The 2024 Nobel Prize in Chemistry recognized Demis Hassabis, John Jumper And David Baker for the usage of machine learning to tackle certainly one of the largest challenges in biology: predicting the 3D shape of proteins and designing them from the bottom up.
This 12 months's award stood out for recognizing research that comes from a technology company: DeepMind, an AI research startup acquired by Google in 2014. Most of the Nobel Prizes in Chemistry thus far have gone to academic researchers. Many award winners then founded startup firms to further expand and commercialize their groundbreaking work – for instance CRISPR gene editing technology And Quantum dots – however the research, from start to complete, was not conducted in a business setting.
Although the Nobel Prizes for Physics and Chemistry are awarded individually, there’s an interesting connection between the winning research in these fields in 2024. The Physics Prize went to 2 computer scientists who laid the inspiration for machine learning, while the chemistry laureates were rewarded for his or her use of machine learning to resolve certainly one of biology's best mysteries: how proteins fold.
The 2024 Nobel Prizes underscore each the importance of such a artificial intelligence and the indisputable fact that science today often crosses traditional boundaries and combines different fields to realize groundbreaking results.
The challenge of protein folding
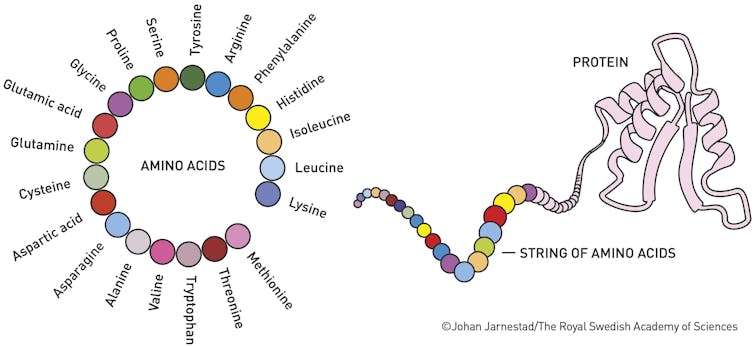
Proteins are the molecular machines of life. They make up a major a part of our body, including muscles, enzymes, hormones, blood, hair and cartilage.
©Johan Jarnestad/Royal Swedish Academy of Sciences
It is significant to know the structures of proteins because their shape determines their functions. In 1972 Christian Anfinsen received the Nobel Prize in chemistry for detecting the sequence of amino acid constructing blocks of a protein determines the form of the proteinwhich in turn affects its function. If a protein folds incorrectly, it might not function properly and might result in diseases equivalent to: Alzheimer, Cystic fibrosis or diabetes.
The overall shape of a protein will depend on the tiny interactions, attractions and repulsions between all of the atoms of the amino acids that compose it. Some wish to be together, some don't. The protein twists and folds into its final shape based on many 1000’s of those chemical interactions.
For many years, certainly one of the best challenges in biology has been predicting the form of a protein based solely on its amino acid sequence. Although researchers can now predict shape, we still don't understand how the proteins maneuver themselves into their specific shapes and minimize the repulsions of all interatomic interactions in a number of microseconds.
To understand how proteins work and stop misfolding, scientists needed a approach to predict how proteins fold, but solving this puzzle was no easy task.
In 2003, he became a biochemist on the University of Washington David Baker wrote Rosettaa pc program for designing proteins. In doing so, he showed that it was possible to reverse the issue of protein folding Designing a protein shape after which predicting it the amino acid sequence needed for production.
It was an exceptional breakthrough, however the form chosen for the calculation was easy and the calculations complex. To routinely design recent proteins with desired structures required a serious paradigm shift.
A brand new era of machine learning
Machine learning is a type of AI by which computers learn to resolve problems by analyzing large amounts of information. It has been utilized in various fields, from Play And Speech recognition To autonomous vehicles And scientific research. The idea behind machine learning is to take advantage of hidden patterns in data to reply complex questions.
This approach took an enormous leap in 2010 when Demis Hassabis was a co-founder DeepMindan organization that goals to mix neuroscience with AI to resolve real-world problems.
Hassabis, a 4-year-old chess prodigy, quickly made headlines AlphaZeroan AI that has taught itself to play chess at a superhuman level. In 2017, AlphaZero significantly beat the world's best computer chess program Stockfish-8. AI's ability to learn from its own gameplay quite than counting on pre-programmed strategies marked a turning point within the AI world.
Soon after, DeepMind applied similar techniques to Go, an ancient board game known for its immense complexity. In 2016, his AI program AlphaGo defeated certainly one of the world's best players, Lee Sedol, in a single widely seen game that amazed tens of millions.

AP Photo/Alastair Grant
In 2016, Hassabis shifted DeepMind's focus to a brand new challenge: the issue of protein folding. Under the direction of John Jumpera chemist with a background in protein science, began the AlphaFold project. The team used a big database of experimentally determined protein structures to coach the AI to learn the principles of protein folding. The result was AlphaFold2, an AI that might predict the 3D structure of proteins based on their amino acid sequences with remarkable accuracy.
This was a serious scientific breakthrough. AlphaFold has since predicted the structures of over 200 million proteins – essentially all of the proteins that scientists have sequenced thus far. The extensive database of protein structures is now freely available and accelerates research in biology, medicine and drug development.
Designer proteins to fight disease
Understanding the folding and performance of proteins is crucial for developing recent drugs. Enzymesa style of protein, act as catalysts in biochemical reactions and might speed up or regulate these processes. To treat diseases equivalent to cancer or diabetes, researchers often goal specific enzymes involved in disease pathways. By predicting the form of a protein, scientists can determine where small molecules—potential drug candidates—might bind to it. This is step one in developing recent drugs.
In 2024, DeepMind was launched AlphaFold3an updated version of the AlphaFold program that not only predicts protein shapes but additionally identifies potential binding sites for small molecules. This advance makes it easier for researchers to develop drugs that focus on exactly the correct proteins.
Google bought Deepmind for supposedly around Half a billion dollars in 2014. Google DeepMind has now launched a brand new company: Isomorphic labsto collaborate with pharmaceutical firms on real-world drug development using these AlphaFold3 predictions.

Ian C. Haydon/UW Medicine Institute for Protein Design
For his part, David Baker has continued to make significant contributions to protein science. His team on the University of Washington developed an AI-based method called “family-wide hallucination“, which they used to develop completely recent proteins from scratch. Hallucinations are recent patterns—on this case, proteins—which are plausible, meaning they fit well with patterns within the AI's training data. These recent proteins included a light-emitting enzyme, showing that machine learning may help develop novel synthetic proteins. These AI tools offer recent opportunities to engineer functional enzymes and other proteins that might never have evolved naturally.
AI will enable the following chapter of research
The Nobel Prize-winning achievements of Hassabis, Jumper and Baker show that machine learning will not be only a tool for computer scientists – it’s now a vital a part of the long run of biology and medicine.
By tackling certainly one of biology's most difficult problems, the 2024 Prize winners have opened up recent possibilities in drug discovery, personalized medicine, and even our understanding of the chemistry of life itself.
image credit : theconversation.com

















Leave a Reply