In the twentieth century when a routine infection was treated with a typical antibioticIt was expected. Over time, the microbes liable for these infections have developed to avoid the medication that they need to remove.
Every yr there may be More than 2.8 million antibiotic-resistant infections within the United States, which results in over 35,000 deaths and 4.6 billion US dollars in health costs of 4.6 billion dollars. If antibiotics change into less effective, antimicrobial resistance is an increasing threat to public health.
The antimicrobial resistance arose within the Nineteen Forties with the rise of a serious threat Penicillin resistance. In the Nineteen Nineties it was too escalated Global problem. Decades later, critical questions remain: How does antimicrobial resistance arise and the way can scientists follow the hidden changes that result in this? Why does the resistance stay undetected in a couple of microbes until an outbreak occurs? Filling these knowledge gaps is crucial to forestall future outbursts, improve treatment results and save lives.
https://www.youtube.com/watch?v=EKYAUG9RSSU
Over the years my work as Microbiologist and biomedical scientist has focused on examining the genetics of infectious microbes. My colleagues and I identified A Resistance gene, which was previously undetected within the USA With the assistance of genetic and arithmetic methods that will help to prove and follow antimicrobial resistance.
Challenges when recognizing resistance
Antimicrobial resistance is a natural process Where microbes consistently become a defense mechanism and gain genetic changes that improve their survival.
Unfortunately, human activities can give you the option to Accelerate this process. The overuse and abuse of antibiotics in healthcare, agriculture and the environment urges bacteria to alter too genetically in a way that allows them to survive the medication they need to kill.
Early proof of antimicrobial resistance is of crucial importance for effective treatment. surveillance As a rule, a laboratory sample of patients with compact infections begins, that are then analyzed to discover potential antimicrobial resistance. This was traditionally used Cultural -based methods This includes exposing microbes within the laboratory and observing whether or not they have survived to find out whether or not they are resistant. In addition to supporting authorities and researchers who monitor the spread of antimicrobial resistance, hospitals use this approach Choose for treatment plans.
However, cultural -based approaches have Some restrictions. Resistant infections often remain unnoticed until the antibiotics fail, which slows down each detection and intervention processes. In addition, recent resistance genes can already escape.
Genomics of antimicrobial resistance
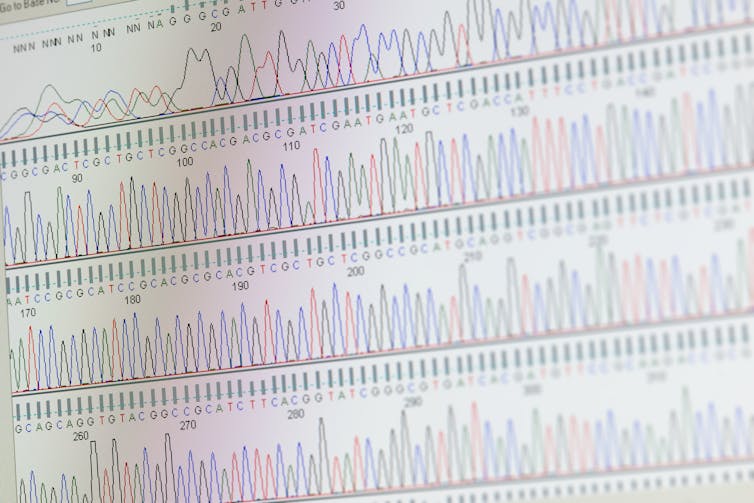
In order to address these challenges, researchers have Integrated genomic sequencing In antimicrobial monitoring. By sequencing the whole genome, we will analyze the whole DNA in a microbial sample as a way to maintain a comprehensive view of all existing genes-an impact on the genes liable for resistance. With the calculation tools of BioinformaticsResearchers can efficiently process large amounts of genetic data as a way to improve the detection of resistance threats.
Despite its benefits, the mixing of genomic sequencing into the monitoring of antimicrobial resistance Present some challenges own. High costs, quality assurance and an absence of trained bioinformatics make it difficult to implement. In addition, the complexity of the interpretation of genomic data can restrict its use in the choice -making technique of clinical and public health.
HH5800/iStock via Getty Images Plus
The definition of international standards could help to make sequencing and bioinformatics in the whole genome a very reliable tool for monitoring resistance. The World Health Organization recommends that Labors are strictly followed Quality control measures To ensure precise and comparable results. This includes the usage of reliable, user -friendly computer tools and approved microbial databases. Additional strategies Inclusion of the investment in training programs and the promotion of the collaborations between hospitals, research laboratories and universities.
Discovery of a resistance gene
My colleagues and I analyzed samples, which were taken from several animal species between 1982 and 1999, which were combined by the whole genome sequencing and bioinformatics. We discovered A Resistance to resistance Called Blasc-1, which has discovered the invention in US cattle base for a long time.
The Blasc-1 gene gives microbes against several critical antibiotics, including Ampicillin, amoxicillin clavulanic acid and to a certain extent, Cephalosporins and carbapenems. These drugs are crucial for the treatment of infections in each humans and animals.
Niaid/FlickrPresent CC BY-SA
The Blasc-1 gene was probably not reported since the routine monitoring normally goals at well-known resistance genes and has overlapping functions with other genes. Gaps in bioinformatics skills may have hindered its identification.
The failure to acknowledge genes like Blasc-1 accuses its potential role of earlier treatment errors. Between 2015 and 2018, the centers for the control and prevention of diseases began implementing the sequencing of the whole genome for Routine monitoring of . Studies carried out during this era 77% of the outbreaks with multi -stage outbursts were related to cattle which might be resistant.
These missed genes have a major impact on food safety and public health. Unknown antimicrobial resistance genes can spread through food animals, contaminated foods, processing environments and agricultural drains, in order that resistant bacteria can still exist and may reach people. These resistant bacteria result in infections which might be harder to treat and increase the danger of outbreaks. In addition, the worldwide movement of individuals, cattle and were that these resistant tribes can easily cross boundariesto remodel local outbreaks into worldwide health threats.
Identifying recent resistance genes not only fulfills a critical knowledge gap, but in addition shows how genomic and arithmetic approaches can recognize hidden resistance mechanisms before they represent widespread threats.
Reinforcement of monitoring
As an antimicrobial resistance, A continues and accepts A A health approach This integrates human, animal and ecological aspects, will help not exceed the flexibility of individuals to combat it.
Initiatives like that Quarter partner Amr Multi-partner Trust Fund Support for programs that strengthen global collaborative surveillance, promote responsible antimicrobial use and promote the event of sustainable alternatives. Make sure that researchers around the globe follow common research standards, more laboratory- especially those in countries with low and medium income- can contribute to contributing to Global monitoring efforts.
The health of future generations is dependent upon the flexibility of the world to make sure food safety and to guard public health at a world level. In the continued struggle between microbial evolution and human innovation, vigilance and adaptableness are of crucial importance as a way to remain prematurely.
image credit : theconversation.com

















Leave a Reply